Welcome to RCM - The Bent Lab LRR Conservation Mapping Program
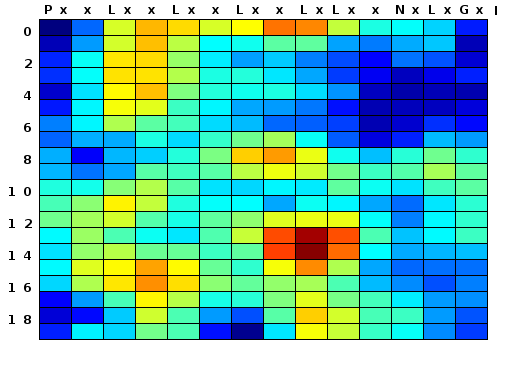 The RCM program predicts biologically functional sites in a leucine-rich repeat (LRR) domain by identifying conserved surface regions on a model of the folded protein. The input is two or more user-entered amino acid sequences, typically from orthologous or paralogous proteins with a similar number of LRR repeats. You can enter just the LRR domain of each protein, or more of the protein (in either case, be prepared to do some editing). The LRR sequence will be rearranged to roughly approximate a folded LRR, and then the program will search iteratively through clusters of predicted adjacent surface-exposed residues and identify a weighted average amino acid conservation score for each cluster. A color map is generated that highlights predicted regions of evolutionary conservation or diversification, which frequently correspond to the key functional sites on the LRR.
The RCM program predicts biologically functional sites in a leucine-rich repeat (LRR) domain by identifying conserved surface regions on a model of the folded protein. The input is two or more user-entered amino acid sequences, typically from orthologous or paralogous proteins with a similar number of LRR repeats. You can enter just the LRR domain of each protein, or more of the protein (in either case, be prepared to do some editing). The LRR sequence will be rearranged to roughly approximate a folded LRR, and then the program will search iteratively through clusters of predicted adjacent surface-exposed residues and identify a weighted average amino acid conservation score for each cluster. A color map is generated that highlights predicted regions of evolutionary conservation or diversification, which frequently correspond to the key functional sites on the LRR.
Note that use of this program requires multiple mouse clicks, and sequence editing, and some knowledge of LRR domains.
 LRR Conservation Mapping
LRR Conservation Mapping